Running workflow
A workflow is a set of tasks whose relationship is described with a directed acyclic graph (DAG). In a DAG, a parent task can be connected to one or more child tasks with each edge directed from the parent task to a child task. A child task processes the output data of the parent task. It is possible to configure each child task to get started when the parent task produces the entire output data or partially produces the output data, depending on the use-case. If a child task is configured as the latter both parent and child tasks will run in parallel for a while, which will reduce the total execution time of the workflow. Currently tasks have to be PanDA tasks, but future versions will support more backend systems such as local batch systems, production system, kubernetes-based resources, and other workflow management systems, to run some tasks very quickly or outsource sub-workflows.
The user describes a workflow using a yaml-based language, Common Workflow Language
(CWL),
or a Python-based language, Snakemake,
and submits it to PanDA using pchain.
This page explains how to use pchain as well as how to describe workflows.
Remark: It could be a bit cumbersome and error-prone to edit CWL or snakemake files. We are developing a handy tool/interface that translates graphical diagrams to workflow descriptions written in python or yaml on behalf of users. However, you have to play with CWL or snakemake files for now.
Workflow examples with CWL
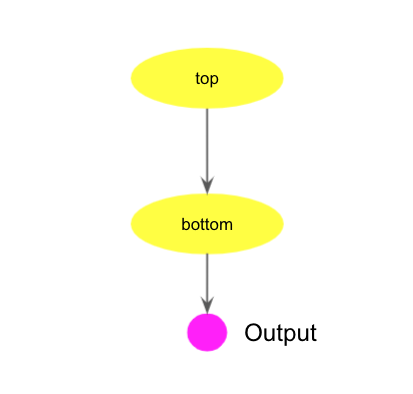
Simple task chain
The following cwl code shows a parent-child chain of two prun tasks.
cwlVersion: v1.0
class: Workflow
inputs: []
outputs:
outDS:
type: string
outputSource: bottom/outDS
steps:
top:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed.txt"
opt_args:
default: "--outputs seed.txt --nJobs 3 --avoidVP"
out: [outDS]
bottom:
run: prun
in:
opt_inDS: top/outDS
opt_exec:
default: "echo %IN > results.root"
opt_args:
default: "--outputs results.root --forceStaged --avoidVP"
out: [outDS]
The class field must be Workflow to indicate this code describes a workflow.
There are two prun tasks in the workflow and defined as top and bottom steps in steps section.
The inputs section is empty since the workflow doesn’t take any input data.
The outputs section describes the output parameter of the workflow, and it takes only one string type parameter
with an arbitrary name.
The outputSource connects the output parameter of the bottom step to the workflow output parameter.
In the steps section, each step represents a task with an arbitrary task name, such as top
and bottom.
The run filed of a prun task is prun. The in section specifies a set of parameters
correspond to command-line options of prun.
Here is a list of parameters in the in section to run a prun task.
Parameter |
Corresponding prun option |
|---|---|
opt_inDS |
—inDS (string) |
opt_inDsType |
No correspondence. Type of inDS (string) |
opt_secondaryDSs |
—secondaryDSs (a list of strings) |
opt_secondaryDsTypes |
No correspondence. Types of secondaryDSs (a string array) |
opt_exec |
—exec (string) |
opt_useAthenaPackages |
—useAthenaPackages (bool) |
opt_containerImage |
—containerImage (string) |
opt_args |
all other prun options except for listed above (string) |
All options opt_xyz except opt_args and opt_xyzDsTypes can be mapped to —xyz of prun.
opt_args specifies all other prun options such as —outputs, —nFilesPerJob,
and —nJobs.
Essentially,
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed.txt"
opt_args:
default: "--outputs seed.txt --nJobs 3"
corresponds to
prun --exec "echo %RNDM:10 > seed.txt" --outputs seed.txt --nJobs 3
%IN in opt_args is expanded to a list of filenames in the input dataset specified in opt_inDS.
It is possible to use %{DSn} in opt_args as a placeholder for the input dataset name.
The out section specifies the task output with an arbitrary string surrendered by brackets.
Note that it is always a single string even if the task produces multiple outputs.
The output of the top task is passed to opt_inDS of the bottom task.
The bottom task starts processing once the top task produces enough output data,
waits if all data currently available has been processed but the top task is still running,
and finishes once all data from the top task is processed.
The user can submit the workflow to PanDA using pchain that is included in panda-client.
First, create a file called simple_chain.cwl containing the cwl code above.
Next, you need to create an empty yaml file since cwl files work with yaml files that describe workflow inputs.
This example doesn’t take an input, so the yaml file can be empty.
$ touch dummy.yaml
$ pchain --cwl simple_chain.cwl --yaml dummy.yaml --outDS user.<your_nickname>.blah
$ touch dummy.yaml
$ pchain --cwl simple_chain.cwl --yaml dummy.yaml --outDS user.<your_nickname>.blah \
--vo wlcg --prodSourceLabel test --workingGroup ${PANDA_AUTH_VO}
pchain automatically sends local *.cwl, *.yaml, and *.json files to PanDA together with the workflow.
--outDS is the basename of the datasets for output and log files. Once the workflow is submitted,
the cwl and yaml files are parsed on the server side to generate tasks
with sequential numbers in the workflow. The system uses a combination of the sequential number
and the task name, such as 000_top and 001_bottom, as a unique identifier for each task.
The actual output dataset name is a combination of --outDS, the unique identifier, and —outputs
in opt_args. For example, the output dataset name of the top task is
user.<your_nickname>.blah_000_top_seed.txt
and that of the bottom is user.<your_nickname>.blah_001_bottom_results.root.
If —outputs is a comma-separate
output list, one dataset is created for each output type.
To see all options of pchain
pchain --helpGroup ALL
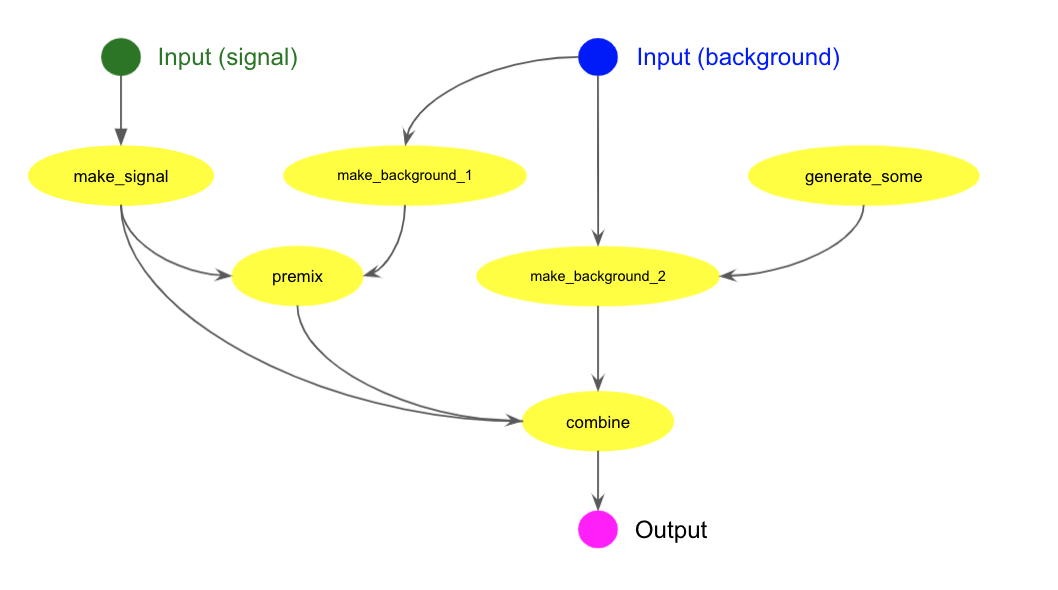
More complicated chain
The following cwl example describes more complicated chain as shown in the picture below.
cwlVersion: v1.0
class: Workflow
requirements:
MultipleInputFeatureRequirement: {}
inputs:
signal: string
background: string
outputs:
outDS:
type: string
outputSource: combine/outDS
steps:
make_signal:
run: prun
in:
opt_inDS: signal
opt_containerImage:
default: docker://busybox
opt_exec:
default: "echo %IN > abc.dat; echo 123 > def.zip"
opt_args:
default: "--outputs abc.dat,def.zip --nFilesPerJob 5"
out: [outDS]
make_background_1:
run: prun
in:
opt_inDS: background
opt_exec:
default: "echo %IN > opq.root; echo %IN > xyz.pool"
opt_args:
default: "--outputs opq.root,xyz.pool --nGBPerJob 10"
out: [outDS]
premix:
run: prun
in:
opt_inDS: make_signal/outDS
opt_inDsType:
default: def.zip
opt_secondaryDSs: [make_background_1/outDS]
opt_secondaryDsTypes:
default: [xyz.pool]
opt_exec:
default: "echo %IN %IN2 > klm.root"
opt_args:
default: "--outputs klm.root --secondaryDSs IN2:2:%{SECDS1}"
out: [outDS]
generate_some:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > gen.root"
opt_args:
default: "--outputs gen.root --nJobs 10"
out: [outDS]
make_background_2:
run: prun
in:
opt_inDS: background
opt_containerImage:
default: docker://alpine
opt_secondaryDSs: [generate_some/outDS]
opt_secondaryDsTypes:
default: [gen.root]
opt_exec:
default: "echo %IN > ooo.root; echo %IN2 > jjj.txt"
opt_args:
default: "--outputs ooo.root,jjj.txt --secondaryDSs IN2:2:%{SECDS1}"
out: [outDS]
combine:
run: prun
in:
opt_inDS: make_signal/outDS
opt_inDsType:
default: abc.dat
opt_secondaryDSs: [premix/outDS, make_background_2/outDS]
opt_secondaryDsTypes:
default: [klm.root, ooo.root]
opt_exec:
default: "echo %IN %IN2 %IN3 > aaa.root"
opt_args:
default: "--outputs aaa.root --secondaryDSs IN2:2:%{SECDS1},IN3:5:%{SECDS2}"
out: [outDS]
The workflow takes two inputs, signal and background. The signal is used as input for
the make_signal
task, while the background is used as input for the make_background_1 and
make_background_2 tasks.
The make_signal task runs in the busybox container as specified in opt_containerImage, to produce two
types of output data, abc.dat and def.zip, as specified in opt_args.
If the parent task produces multiple types of output data and the child task uses some of them,
their types need to be specified in opt_inDsType.
The premix task takes def.zip from the make_signal task and xyz.pool
from the make_background_1 task.
Output data of parent tasks can be passed to a child task as secondary inputs. In this case, they are
specified in opt_secondaryDSs and their types are specified in opt_secondaryDsTypes.
Note that the stream name, the number of files per job, etc, for each secondary input are specified
using —secondaryDSs in opt_args where %{SECDSn} can
be used as a placeholder for the n-th secondary dataset name.
MultipleInputFeatureRequirement is required if opt_secondaryDsTypes take multiple input data.
The workflow inputs are described in a yaml file. E.g.,
cat inputs.yaml
signal: mc16_valid:mc16_valid.900248.PG_singlepion_flatPt2to50.simul.HITS.e8312_s3238_tid26378578_00
background: mc16_5TeV.361238.Pythia8EvtGen_A3NNPDF23LO_minbias_inelastic_low.merge.HITS.e6446_s3238_s3250/
Then submit the workflow.
pchain --cwl sig_bg_comb.cwl --yaml inputs.yaml --outDS user.<your_nickname>.blah
If you need to run the workflow with different input data it enough to submit it with a different yaml file.
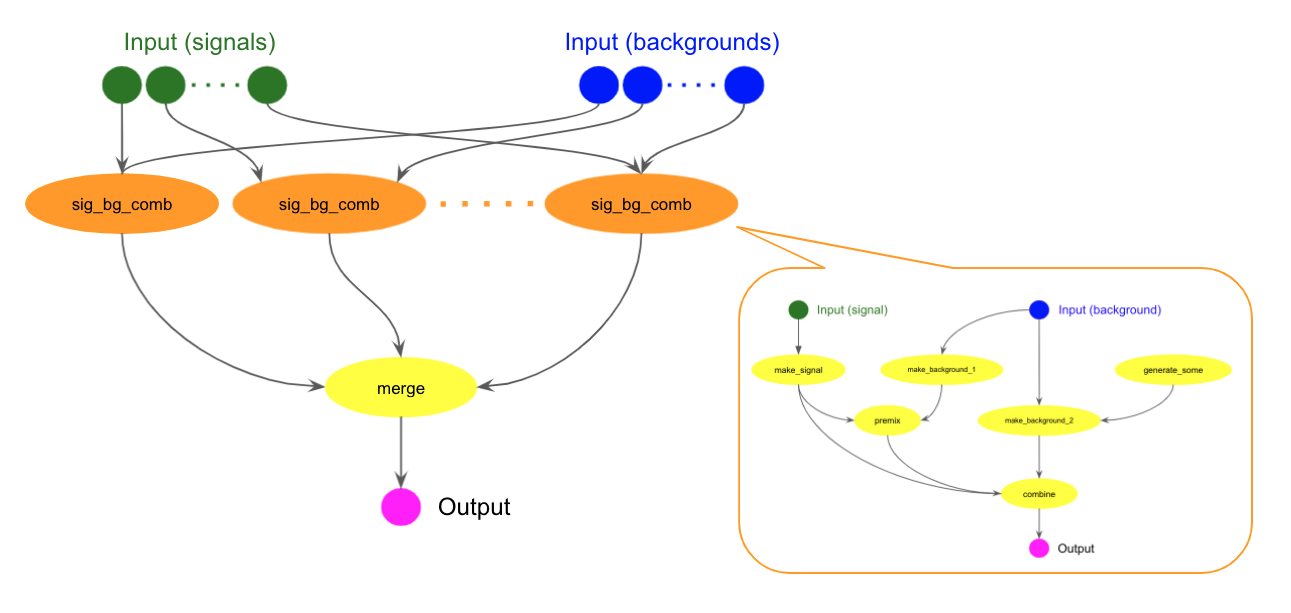
Sub-workflow and parallel execution with scatter
A workflow can be used as a step in another workflow. The following cwl example uses the above sig_bg_comb.cwl in the many_sig_bg_comb step.
cwlVersion: v1.0
class: Workflow
requirements:
ScatterFeatureRequirement: {}
SubworkflowFeatureRequirement: {}
inputs:
signals: string[]
backgrounds: string[]
outputs:
outDS:
type: string
outputSource: merge/outDS
steps:
many_sig_bg_comb:
run: sig_bg_comb.cwl
scatter: [signal, background]
scatterMethod: dotproduct
in:
signal: signals
background: backgrounds
out: [outDS]
merge:
run: prun
in:
opt_inDS: many_sig_bg_comb/outDS
opt_exec:
default: "python merge.py --type aaa --level 3 %IN"
opt_args:
default: "--outputs merged.root"
out: [outDS]
Note that sub-workflows require SubworkflowFeatureRequirement.
It is possible to run a task or sub-workflow multiple times over a list of inputs using
ScatterFeatureRequirement.
A popular use-case is to perform the same analysis step on different samples in a single workflow.
The step takes the input(s) as an array and will run on each element of the array as if it were a single input.
The many_sig_bg_comb step above takes two string arrays, signals and backgrounds,
and specifies in the scatter field that it loops over those arrays.
Output data from all many_sig_bg_comb tasks are fed into the merge task to produce the final output.
The workflow inputs are string arrays like
cat inputs2.yaml
signals:
- mc16_valid:mc16_valid.900248.PG_singlepion_flatPt2to50.simul.HITS.e8312_s3238_tid26378578_00
- valid1.427080.Pythia8EvtGen_A14NNPDF23LO_flatpT_Zprime.simul.HITS.e5362_s3718_tid26356243_00
background:
- mc16_5TeV.361238.Pythia8EvtGen_A3NNPDF23LO_minbias_inelastic_low.merge.HITS.e6446_s3238_s3250/
- mc16_5TeV:mc16_5TeV.361239.Pythia8EvtGen_A3NNPDF23LO_minbias_inelastic_high.merge.HITS.e6446_s3238_s3250/
Then submit the workflow.
pchain --cwl merge_many.cwl --yaml inputs2.yaml --outDS user.<your_nickname>.blah
Using Athena
One or more tasks in a single workflow can use Athena as shown in the example below.
cwlVersion: v1.0
class: Workflow
inputs: []
outputs:
outDS:
type: string
outputSource: third/outDS
steps:
first:
run: prun
in:
opt_exec:
default: "Gen_tf.py --maxEvents=1000 --skipEvents=0 --ecmEnergy=5020 --firstEvent=1 --jobConfig=860059 --outputEVNTFile=evnt.pool.root --randomSeed=4 --runNumber=860059 --AMITag=e8201"
opt_args:
default: "--outputs evnt.pool.root --nJobs 3"
opt_useAthenaPackages:
default: true
out: [outDS]
second:
run: prun
in:
opt_inDS: first/outDS
opt_exec:
default: "echo %IN > results.txt"
opt_args:
default: "--outputs results.txt"
out: [outDS]
third:
run: prun
in:
opt_inDS: second/outDS
opt_exec:
default: "echo %IN > poststep.txt"
opt_args:
default: "--outputs poststep.txt --athenaTag AnalysisBase,21.2.167"
opt_useAthenaPackages:
default: true
out: [outDS]
opt_useAthenaPackages corresponds to --useAthenaPackages of prun to remotely setup Athena with your
locally-built packages.
You can use a different Athena version by specifying —athenaTag in opt_args.
To submit the task, first you need to setup Athena on local computer, and execute pchain
with --useAthenaPackages that automatically collect various Athena-related information
from environment variables and uploads a sandbox file from your locally-built packages.
pchain --cwl athena.cwl --yaml inputs.yaml --outDS user.<your_nickname>.blah --useAthenaPackages
Using Conda
You can build a container image to setup a conda environment and use it in a workflow. The following Dockerfile installs Miniconda and creates an environment with PyROOT and PyYAML.
FROM docker.io/almalinux:9
RUN yum update -y
RUN yum install -y wget
RUN mkdir -p /opt/miniconda3 && \
wget https://repo.anaconda.com/miniconda/Miniconda3-latest-Linux-x86_64.sh -O /tmp/miniconda.sh && \
bash /tmp/miniconda.sh -b -u -p /opt/miniconda3 && \
rm -rf /tmp/miniconda.sh
RUN adduser auser
RUN groupadd zp
RUN usermod -a -G zp auser
USER auser
RUN /opt/miniconda3/bin/conda init bash && . ~/.bashrc && conda create -n testenv -y -c conda-forge root pyyaml
RUN yum clean all
CMD ["/bin/bash"]
You need to upload your container image to a container registry, such as GitLab container registry,
and specify the registry path in opt_containerImage as shown in the example below.
cwlVersion: v1.0
class: Workflow
inputs: []
outputs:
outDS:
type: string
outputSource: bottom/outDS
steps:
top:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed.txt"
opt_args:
default: "--outputs seed.txt --nJobs 3 --avoidVP"
out: [outDS]
bottom:
run: prun
in:
opt_inDS: top/outDS
opt_exec:
default: "export HOME=/home/auser; . /home/auser/.bashrc; conda activate testenv; root --version; ls -la"
opt_args:
default: "--forceStaged --avoidVP"
opt_containerImage:
default: "docker://ghcr.io/someone/test-actions:main"
out: [outDS]
As specified in opt_exec, the bottom task first initializes and activates the conda environment created
in the build step of the container image, and then executes root and ls as a demonstration.
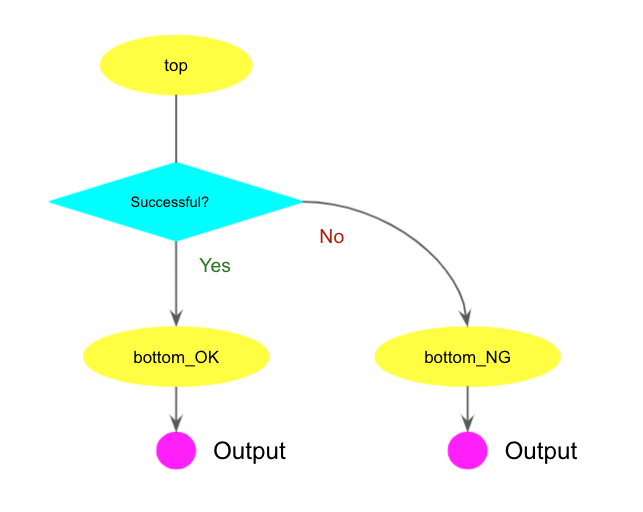
Conditional workflow
Workflows can contain conditional steps executed based on their input. This allows workflows
to wait execution of subsequent tasks until previous tasks are done, and
to skip subsequent tasks based on results of previous tasks.
The following example contains conditional branching based on the result of the first step.
Note that this workflows conditional branching require InlineJavascriptRequirement and CWL version 1.2 or higher.
cwlVersion: v1.2
class: Workflow
requirements:
InlineJavascriptRequirement: {}
inputs: []
outputs:
outDS_OK:
type: string
outputSource: bottom_OK/outDS
outDS_NG:
type: string
outputSource: bottom_NG/outDS
steps:
top:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed.txt"
opt_args:
default: "--outputs seed.txt --nJobs 2"
out: [outDS]
bottom_OK:
run: prun
in:
opt_inDS: top/outDS
opt_exec:
default: "echo %IN > results.txt"
opt_args:
default: "--outputs results.txt --forceStaged"
out: [outDS]
when: $(self.opt_inDS)
bottom_NG:
run: prun
in:
opt_inDS: top/outDS
opt_exec:
default: "echo hard luck > bad.txt"
opt_args:
default: "--outputs bad.txt"
out: [outDS]
when: $(!self.opt_inDS)
Both bottom_OK and bottom_NG steps take output data of the top step as input.
The new property when specifies the condition validation expression that is interpreted by JavaScript.
self.blah in the expression represents the input parameter blah of the step that is connected
to output data of the parent step. If the parent step is successful self.blah gives True
while !self.blah gives False. It is possible to create more complicated expressions using
logical operators (&& for AND and || for OR) and parentheses. The step is executed when the
whole expression gives True.
The bottom_NG step is executed when the top step fails and $(!self.opt_inDS) gives
True. Note that in this case output data from the top step is empty and
the prun task in the bottom_NG step is executed without --inDS.
Involving hyperparameter optimization
It is possible to run Hyperparameter Optimization (HPO) and chain it with other tasks in the workflow. The following example shows a chain of HPO and prun tasks.
cwlVersion: v1.0
class: Workflow
inputs: []
outputs:
outDS:
type: string
outputSource: post_proc/outDS
steps:
pre_proc:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed.txt"
opt_args:
default: "--outputs seed.txt --nJobs 2 --avoidVP"
out: [outDS]
main_hpo:
run: phpo
in:
opt_trainingDS: pre_proc/outDS
opt_args:
default: "--loadJson conf.json"
out: [outDS]
when: $(self.opt_trainingDS)
post_proc:
run: prun
in:
opt_inDS: main_hpo/outDS
opt_exec:
default: "echo %IN > anal.txt"
opt_args:
default: "--outputs anal.txt --nFilesPerJob 100 --avoidVP"
out: [outDS]
when: $(self.opt_inDS)
where the output data of the pre_proc step is used as the training data for the main_hpo step,
and the output data metrics.tgz of the main_hpo step is used as the input for the
post_proc step.
Both main_hpo and post_proc steps specify when since they waits until the upstream step is done.
The run filed of a phpo task is phpo.
Here is a list of parameters in the in section to run a prun task.
Parameter |
Corresponding phpo option |
|---|---|
opt_trainingDS |
—trainingDS (string) |
opt_trainingDsType |
No correspondence. Type of trainingDS (string) |
opt_args |
all other phpo options except for listed above (string) |
opt_trainingDS can be omitted if the HPO task doesn’t take a training dataset.
Note that you can put most phpo options in a json and specify the json filename in —loadJson
in opt_args, rather than constructing a complicated string in opt_args.
cat config.json
{
"evaluationContainer": "docker://gitlab-registry.cern.ch/zhangruihpc/evaluationcontainer:mlflow",
"evaluationExec": "bash ./exec_in_container.sh",
"evaluationMetrics": "metrics.tgz",
"steeringExec": "run --rm -v \"$(pwd)\":/HPOiDDS gitlab-registry.cern.ch/zhangruihpc/steeringcontainer:0.0.1 /bin/bash -c \"hpogrid generate --n_point=%NUM_POINTS --max_point=%MAX_POINTS --infile=/HPOiDDS/%IN --outfile=/HPOiDDS/%OUT -l nevergrad\""
}
pchain -cwl hpo.cwl --yaml dummy.yaml --outDS user.<your_nickname>.blah
Loops in workflows
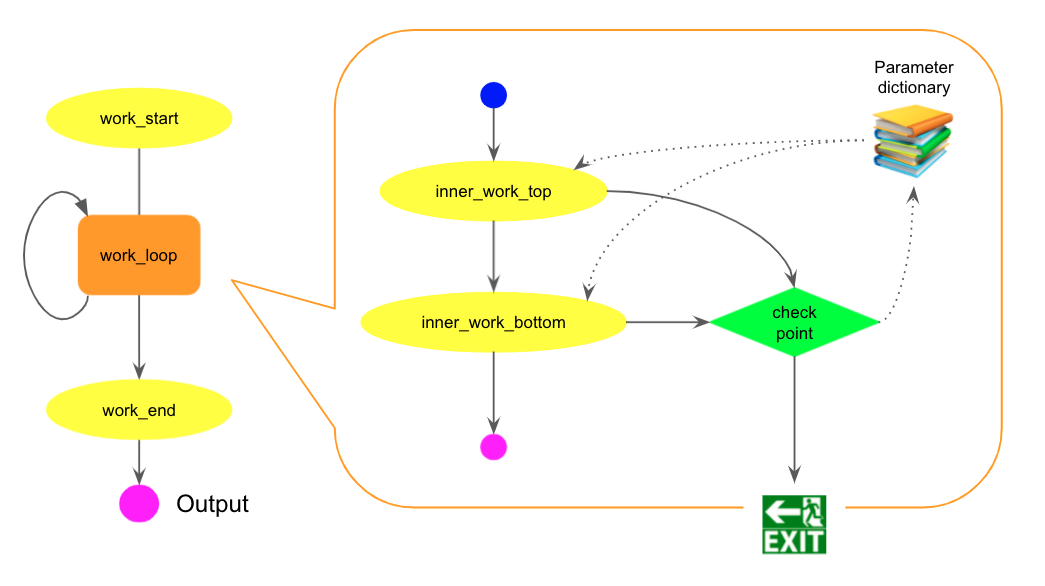
Users can have loops in their workflows. Each loop is represented as a sub-workflow with a parameter dictionary. All tasks in the sub-workflow share the dictionary to generate actual steps. There is a special type of tasks, called junction, which read outputs from upstream tasks, and produce json files to update the parameter dictionary and/or make a decision to exit from the sub-workflow. The sub-workflow is iterated until one of junctions decides to exit. The new iteration is executed with the updated values in the parameter dictionary, so that each iteration can bring different results.
The run filed of a junction is junction.
Essentially, a junction is a simplified prun task that processes all input files in a single job
to produce a json file, so there are only a few parameters in the in section of a junction, as shown below.
Parameter |
Corresponding prun option |
|---|---|
opt_inDS |
Input datasets (a list of strings) |
opt_inDsType |
Types of input datasets (a list of strings. optional) |
opt_exec |
The execution string |
opt_containerImage |
Container image name (string. optional) |
opt_args |
To define additional output files in –outputs |
For example, the following pseudo-code snippet has a single loop
out1 = work_start()
xxx = 123
yyy = 0.456
while True:
out2 = inner_work_top(out1, xxx)
out3 = inner_work_bottom(out2, yyy)
xxx = out2 + 1
yyy *= out3
if yyy > xxx:
break
out4 = work_end(out3)
The code is described using CWL as follows.
cwlVersion: v1.0
class: Workflow
requirements:
SubworkflowFeatureRequirement: {}
inputs: []
outputs:
outDS:
type: string
outputSource: work_end/outDS
steps:
work_start:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed.txt"
opt_args:
default: "--outputs seed.txt --nJobs 2 --avoidVP"
out: [outDS]
work_loop:
run: loop_body.cwl
in:
dataset: work_start/outDS
out: [outDS]
hints:
- loop
work_end:
run: prun
in:
opt_inDS: work_loop/outDS
opt_exec:
default: "echo %IN > results.root"
opt_args:
default: "--outputs results.root --forceStaged"
out: [outDS]
The work_loop step describes the looping stuff in the while block of the pseudo-code snippet.
It runs a separate CWL file loop_body.cwl and
has loop in the hints section to iterate.
cwlVersion: v1.0
class: Workflow
inputs:
dataset:
type: string
param_xxx:
type: int
default: 123
param_yyy:
type: float
default: 0.456
outputs:
outDS:
type: string
outputSource: inner_work_bottom/outDS
steps:
inner_work_top:
run: prun
in:
opt_inDS: dataset
opt_exec:
default: "echo %IN %{xxx} %{i} > seed.txt"
opt_args:
default: "--outputs seed.txt --avoidVP"
out: [outDS]
inner_work_bottom:
run: prun
in:
opt_inDS: inner_work_top/outDS
opt_exec:
default: "echo %IN %{yyy} > results.root"
opt_args:
default: "--outputs results.root --forceStaged"
out: [outDS]
checkpoint:
run: junction
in:
opt_inDS:
- inner_work_top/outDS
- inner_work_bottom/outDS
opt_exec:
default: "echo %{DS1} %{DS2} aaaa; echo '{\"x\": 456, \"to_continue\": false}' > results.json"
out: []
The local variables in the loop like xxx and yyy are defined in the inputs section
of loop_body.cwl with the param_ prefix and their initial values. They are internally
translated to a parameter dictionary shared by all tasks in the sub-workflow.
In the execution, %{blah} in opt_args is replaced with the actual value in the dictionary.
A loop count is incremented for each iteration and is inserted to the output dataset names, like
user.<your_nickname>.blah_<loop_count>_<output>,
so tasks always produce unique output datasets.
It is possible to specify the loop count explicitly in opt_exec using %{i}.
In other words, param_i is reserved and thus cannot be used as user’s local variable.
In each iteration, the checkpoint step runs a junction to read outputs from inner_work_top
and inner_work_bottom steps, and produce a json file to update values in the parameter dictionary
and/or make a decision to exit from the loop.
The actual dataset names are passed to the execution string through placeholders, %{DSn}
in opt_exec, which represents the n-th dataset name.
The json filename must be results.json.
The file contains key-values to update the local variables in the parameter dictionary.
It can also contain a special key-value, to_continue: True, to repeat the loop.
If to_continue is False, the key-value is missing, or results.json is not produced,
the workflow exits from the loop and proceeds to subsequent steps outside of the loop.
It is possible to specify additional output files via —outputs in opt_args if the junction step
produces other output files in addition to results.json. Subsequent steps can use those output files
as input.
Loop + scatter
A loop is sequential iteration of a sub-workflow, while a scatter is a horizontal parallelization of independent sub-workflows. They can be combined to describe complex workflows.
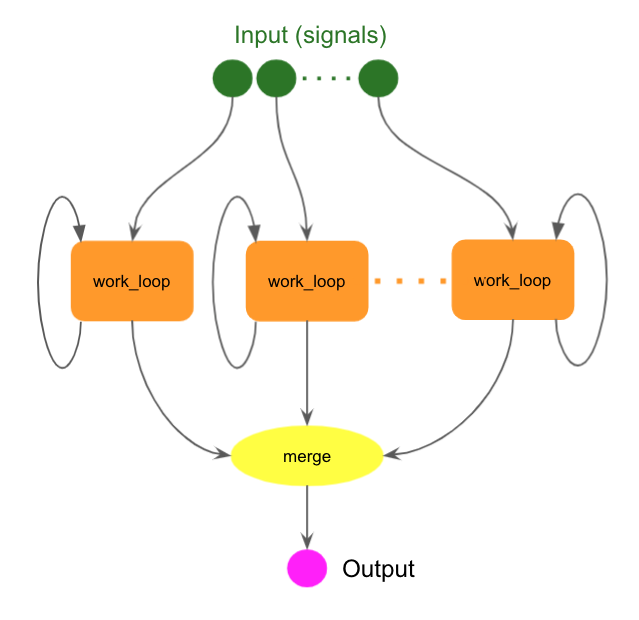
Running multiple loops in parallel
The following example runs multiple loops in parallel.
cwlVersion: v1.0
class: Workflow
requirements:
ScatterFeatureRequirement: {}
SubworkflowFeatureRequirement: {}
inputs:
signals: string[]
outputs:
outDS:
type: string
outputSource: work_end/outDS
steps:
work_loop:
run: loop_body.cwl
scatter: [dataset]
scatterMethod: dotproduct
in:
dataset: signals
out: [outDS]
hints:
- loop
merge:
run: prun
in:
opt_inDS: work_loop/outDS
opt_exec:
default: "echo %IN > results.root"
opt_args:
default: "--outputs results.root --forceStaged"
out: [outDS]
The work_loop step has the loop hint and is scattered over the list of inputs.
Parallel execution of multiple sub-workflows in a single loop
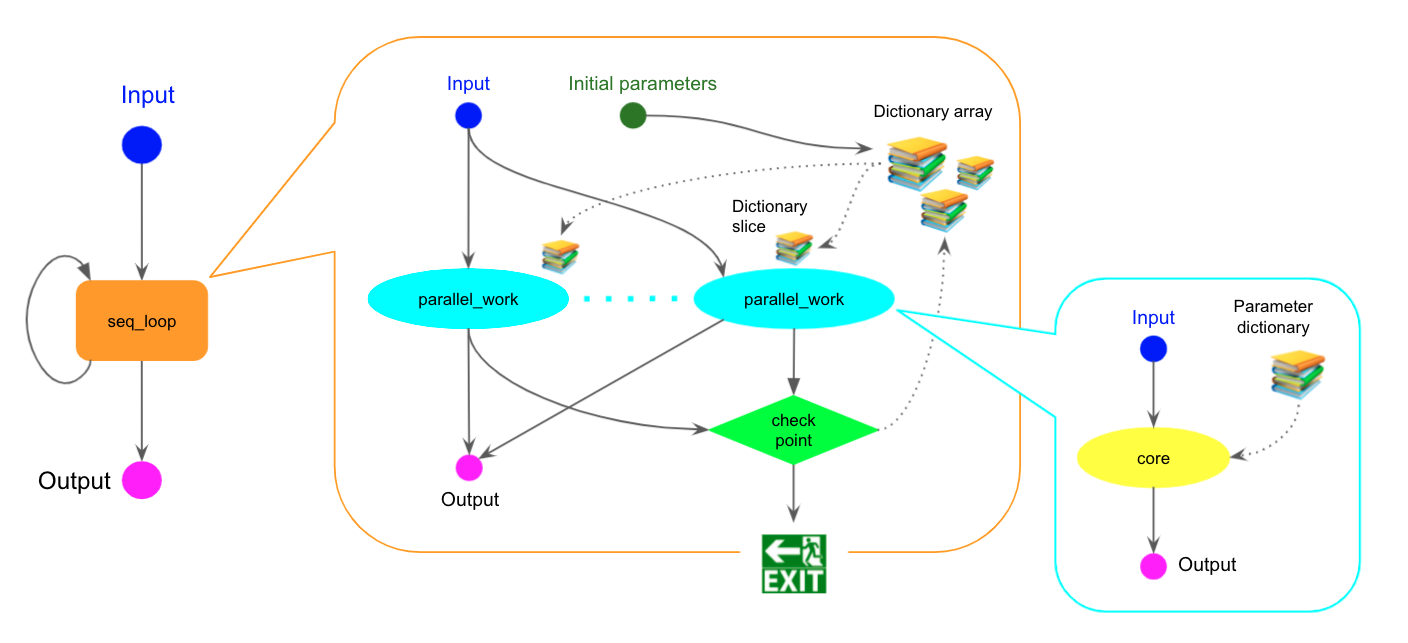
Here is another example of the loop+scatter combination that has a single loop where multiple sub-workflows are executed in parallel.
cwlVersion: v1.0
class: Workflow
requirements:
ScatterFeatureRequirement: {}
SubworkflowFeatureRequirement: {}
inputs:
signal: string
outputs:
outDS:
type: string
outputSource: seq_loop/outDS
steps:
seq_loop:
run: scatter_body.cwl
in:
dataset: signal
out: [outDS]
hints:
- loop
The seq_loop step iterates scatter_body.cwl which defines arrays of parameters with the the param_ prefix and initial values; param_xxx and param_yyy. The parallel_work step is scattered over the arrays. The arrays are vertically sliced so that each execution of loop_main.cwl gets only one parameter set. The checkpoint step takes all outputs from the parallel_work step to update the arrays and make a decision to continue the loop.
cwlVersion: v1.0
class: Workflow
requirements:
ScatterFeatureRequirement: {}
SubworkflowFeatureRequirement: {}
inputs:
dataset: string
param_xxx:
type: int[]
default: [123, 456]
param_yyy:
type: float[]
default: [0.456, 0.866]
outputs:
outDS:
type: string
outputSource: /outDS
steps:
parallel_work:
run: loop_main.cwl
scatter: [param_sliced_x, param_sliced_y]
scatterMethod: dotproduct
in:
dataset: dataset
param_sliced_x: param_xxx
param_sliced_y: param_yyy
out: [outDS]
checkpoint:
run: junction
in:
opt_inDS: parallel_work/outDS
opt_exec:
default: " echo '{\"xxx\": [345, 678], \"yyy\": [0.321, 0.567], \"to_continue\": false}' > results.json"
out: []
The sliced values are passed to loop_main.cwl as local variables
param_sliced_x and param_sliced_y in the sub-workflow.
In each iteration the checkpoint step above updates the values in the arrays,
so that %{blah} in opt_args is replaced with the updated value when the task is actually executed.
cwlVersion: v1.0
class: Workflow
inputs:
dataset:
type: string
param_sliced_x:
type: int
param_sliced_y:
type: float
outputs:
outDS:
type: string
outputSource: core/outDS
steps:
core:
run: prun
in:
opt_inDS: dataset
opt_exec:
default: "echo %IN %{sliced_x} %{sliced_y} > seed.txt"
opt_args:
default: "--outputs seed.txt --avoidVP"
out: [outDS]
Using REANA
The following example offloads a part of workflow to REANA.
cwlVersion: v1.0
class: Workflow
inputs: []
outputs:
outDS:
type: string
outputSource: drai/outDS
steps:
ain:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed_ain.txt"
opt_args:
default: "--outputs seed_ain.txt --avoidVP --nJobs 2"
out: [outDS]
twai:
run: prun
in:
opt_exec:
default: "echo %RNDM:10 > seed_twai.txt"
opt_args:
default: "--outputs seed_twai.txt --avoidVP --nJobs 3"
out: [outDS]
drai:
run: reana
in:
opt_inDS:
- ain/outDS
- twai/outDS
opt_exec:
default: "echo %{DS1} %{DS2} > results.root"
opt_args:
default: "--outputs results.root"
opt_containerImage:
default: docker://busybox
out: [outDS]
The ain and twai steps are executed on PanDA, while the drai step reads outputs from
those steps and produce the final output on REANA.
The run filed of a REANA step is reana.
A REANA step is a simplified prun task composed of a single job that
tells input dataset names to the payload through the execution string.
The payload dynamically customizes the sub-workflow description to processes files in those datasets,
submit it to REANA using secrets,
and downloads the results from REANA.
Similarly to junctions, there are only a few parameters in the in section of a REANA step, as shown below.
Parameter |
Corresponding prun option |
|---|---|
opt_inDS |
Input datasets (a list of strings) |
opt_inDsType |
Types of input datasets (a list of strings. optional) |
opt_exec |
The execution string |
opt_containerImage |
Container image name (string. optional) |
opt_args |
all other prun options except for listed above (string) |
The actual dataset names are passed to the execution string through placeholders, %{DSn}
in opt_exec, which represents the n-th dataset name. Note that the container image in opt_containerImage
submits the sub-workflow description to REANA, so it is generally not the container image that processes input files.
REANA steps are internally executed as prun tasks in PanDA, so that all prun options can be specified in opt_args.
Using Gitlab CI Pipelines
It is possible to integrate Gitlab CI pipelines in workflows. The following example executes the un step on PanDA, and then feeds the output to the deux step to trigger a Gitlab CI pipeline.
cwlVersion: v1.0
class: Workflow
inputs: []
outputs:
outDS:
type: string
outputSource: deux/outDS
steps:
un:
run: prun
in:
opt_exec:
default: "echo 1 > out.txt"
opt_args:
default: "--outputs out.txt"
out: [outDS]
deux:
run: gitlab
in:
opt_inDS:
- un/outDS
opt_site:
default: "CERN"
opt_ref:
default: "master"
opt_api:
default: "https://gitlab.cern.ch/api/v4/projects"
opt_projectID:
default: 165337
opt_triggerToken:
default: "MY_T_TOKEN"
opt_accessToken:
default: "MY_A_TOKEN"
opt_input_location:
default: "/eos/atlas/atlascerngroupdisk/blah"
out: [outDS]
A Gitlab CI step, defined with gitlab in the run filed, specifies the following parameters
in the in section.
Parameter |
Description |
|---|---|
opt_inDS |
Input datasets (a list of strings) |
opt_input_location |
The directory name where input files are downloaded (string. optional) |
opt_site |
The site name where the Gitlab CI is reachable (string) |
opt_api |
The API of the Gitlab projects (string, e.g. https://<hostname>/api/v4/projects) |
opt_projectID |
The project ID of the gitlab repository where pipelines run (int) |
opt_ref |
The branch name in the repository (string, e.g. master) |
opt_triggerToken |
The key of the trigger token uploaded to PanDA secrets (string) |
opt_accessToken |
The key of the access token uploaded to PanDA secrets (string) |
To create trigger and access tokens, see Gitlab documentation pages;
how to create a trigger token
and how to create an access token.
Access tokens require only the read_api scope with the Guest role.
Note that trigger and access tokens need to be uploaded to PanDA secrets
with keys specified in opt_triggerToken and ``opt_accessToken, before submitting workflows.
Workflow examples with Snakemake
Snakemake allows users to describe workflows with Python language syntax and JSON configuration files.
Common JSON configuration file for following examples
{
"output": "outDS",
"signal": "mc16_valid:mc16_valid.900248.PG_singlepion_flatPt2to50.simul.HITS.e8312_s3238_tid26378578_00",
"background": "mc16_5TeV.361238.Pythia8EvtGen_A3NNPDF23LO_minbias_inelastic_low.merge.HITS.e6446_s3238_s3250/",
"signals": [
"mc16_valid:mc16_valid.900248.PG_singlepion_flatPt2to50.simul.HITS.e8312_s3238_tid26378578_00",
"valid1.427080.Pythia8EvtGen_A14NNPDF23LO_flatpT_Zprime.simul.HITS.e5362_s3718_tid26356243_00"
],
"backgrounds": [
"mc16_5TeV.361238.Pythia8EvtGen_A3NNPDF23LO_minbias_inelastic_low.merge.HITS.e6446_s3238_s3250/",
"mc16_5TeV:mc16_5TeV.361239.Pythia8EvtGen_A3NNPDF23LO_minbias_inelastic_high.merge.HITS.e6446_s3238_s3250/"
]
}
Simple task chain
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection SmkRuleRedeclaration
rule all:
input: config["output"]
# noinspection SmkRuleRedeclaration
rule top:
params:
opt_exec="echo %RNDM:10 > seed.txt",
opt_args="--outputs seed.txt --nJobs 3 --avoidVP"
output: f"{config['output']}.top"
shell: "prun"
# noinspection SmkRuleRedeclaration
rule bottom:
params:
opt_inDS=rules.top.output[0],
opt_exec="echo %IN > results.root",
opt_args="--outputs results.root --forceStaged --avoidVP"
input: rules.top.output
output: config["output"]
shell: "prun"
# noinspection directives can be used if a workflow is described using PyCharm IDE with SnakeCharm plugin.
In other case these directives can be omitted.
To use parent output as input the user should specify rules.${parent_rule_name}.output[0] as value
for the corresponding parameter (opt_inDS) and specify rules.${parent_rule_name}.output for input keyword.
More complicated chain
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of, param_exp
# noinspection SmkRuleRedeclaration
rule all:
params:
signal=config["signal"],
background=config["background"]
input: config["output"]
rule make_signal:
params:
opt_inDS=param_of("signal",source=rules.all),
opt_containerImage="docker://busybox",
opt_exec="echo %IN > abc.dat; echo 123 > def.zip",
opt_args="--outputs abc.dat,def.zip --nFilesPerJob 5"
output: f"{config['output']}.signal"
shell: "prun"
rule make_background_1:
params:
opt_inDS=param_of("background",source=rules.all),
opt_exec="echo %IN > opq.root; echo %IN > xyz.pool",
opt_args="--outputs opq.root,xyz.pool --nGBPerJob 10"
output: f"{config['output']}.background"
shell: "prun"
rule premix:
params:
opt_inDS=rules.make_signal.output[0],
opt_inDsType="def.zip",
opt_secondaryDSs=[rules.make_background_1.output[0]],
opt_secondaryDsTypes=["xyz.pool"],
opt_exec="echo %IN %IN2 > klm.root",
opt_args=param_exp("--outputs klm.root --secondaryDSs IN2:2:%{SECDS1}")
input: rules.make_signal.output,rules.make_background_1.output
output: f"{config['output']}.premix"
shell: "prun"
rule generate_some:
params:
opt_exec="echo %RNDM:10 > gen.root",
opt_args="--outputs gen.root --nJobs 10"
output: f"{config['output']}.generate_some"
shell: "prun"
rule make_background_2:
params:
opt_inDS=param_of("background",source=rules.all),
opt_containerImage="docker://alpine",
opt_secondaryDSs=[rules.generate_some.output[0]],
opt_secondaryDsTypes=["gen.root"],
opt_exec="echo %IN > ooo.root; echo %IN2 > jjj.txt",
opt_args=param_exp("--outputs ooo.root,jjj.txt --secondaryDSs IN2:2:%{SECDS1}")
input: rules.generate_some.output
output: f"{config['output']}.background_2"
shell: "prun"
rule combine:
params:
opt_inDS=rules.make_signal.output[0],
opt_inDsType="abc.dat",
opt_secondaryDSs=[rules.premix.output[0], rules.make_background_2.output[0]],
opt_secondaryDsTypes=["klm.root", "ooo.root"],
opt_exec="echo %IN %IN2 %IN3 > aaa.root",
opt_args=param_exp("--outputs aaa.root --secondaryDSs IN2:2:%{SECDS1},IN3:5:%{SECDS2}")
input: rules.make_signal.output,rules.premix.output,rules.make_background_2.output
output: config["output"]
shell: "prun"
To use parameter from another step the user should use the helper function param_of from
pandaserver.workflow.snakeparser.utils library.
For example, param_of("${name_of_parameter}",source=rules.all), where ${name_of_parameter} should be replaced by
actual parameter name and source refers to the parameter step.
If step parameter value contains patterns like %{SECDS1} the helper function param_exp should be used.
To use multiple inputs (multiple parents) input keyword should contain all references separated by commas.
Sub-workflow and parallel execution with scatter
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of
# noinspection SmkRuleRedeclaration
rule all:
params:
signals=config["signals"],
backgrounds=config["backgrounds"]
input: config["output"]
# noinspection SmkUnrecognizedSection
rule many_sig_bg_comb:
params:
signal=param_of("signals",source=rules.all),
background=param_of("backgrounds",source=rules.all)
scatter: [param_of("signal"), param_of("background")]
output: f"{config['output']}.many_sig_bg_comb"
shell: "../sig_bg_comb/Snakefile"
# noinspection SmkRuleRedeclaration
rule merge:
params:
opt_inDS=rules.many_sig_bg_comb.output[0],
opt_exec="python merge.py --type aaa --level 3 %IN",
opt_args="--outputs merged.root"
input: rules.many_sig_bg_comb.output
output: config["output"]
shell: "prun"
Using Athena
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection SmkRuleRedeclaration
rule all:
input: config["output"]
rule first:
params:
opt_exec="Gen_tf.py --maxEvents=1000 --skipEvents=0 --ecmEnergy=5020 --firstEvent=1 --jobConfig=860059 " +
"--outputEVNTFile=evnt.pool.root --randomSeed=4 --runNumber=860059 --AMITag=e8201",
opt_args="--outputs evnt.pool.root --nJobs 3",
opt_useAthenaPackages=True
output: f"{config['output']}.first"
shell: "prun"
rule second:
params:
opt_inDS=rules.first.output[0],
opt_exec="echo %IN > results.txt",
opt_args="--outputs results.txt"
input: rules.first.output
output: f"{config['output']}.second"
shell: "prun"
rule third:
params:
opt_inDS=rules.second.output[0],
opt_exec="echo %IN > poststep.txt",
opt_args="--outputs poststep.txt --athenaTag AnalysisBase,21.2.167",
opt_useAthenaPackages=True
input: rules.second.output
output: config["output"]
shell: "prun"
Conditional workflow
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of
# noinspection SmkRuleRedeclaration
rule all:
input: f"{config['output']}.bottom_ok",f"{config['output']}.bottom_ng"
# noinspection SmkRuleRedeclaration
rule top:
params:
opt_exec="echo %RNDM:10 > seed.txt",
opt_args="--outputs seed.txt --nJobs 2"
output: f"{config['output']}.top"
shell: "prun"
# noinspection SmkRuleRedeclaration, SmkUnrecognizedSection
rule bottom_OK:
params:
opt_inDS=rules.top.output[0],
opt_exec="echo %IN > results.txt",
opt_args="--outputs results.txt --forceStaged"
input: rules.top.output
output: f"{config['output']}.bottom_ok"
when: str(param_of("opt_inDS"))
shell: "prun"
# noinspection SmkRuleRedeclaration, SmkUnrecognizedSection
rule bottom_NG:
params:
opt_inDS=rules.top.output[0],
opt_exec="echo hard luck > bad.txt",
opt_args="--outputs bad.txt"
input: rules.top.output
output: f"{config['output']}.bottom_ng"
when: f"!{param_of('opt_inDS')}"
shell: "prun"
when keyword was added as Snakemake language extension to define step conditions.
Involving hyperparameter optimization
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of
# noinspection SmkRuleRedeclaration
rule all:
input: config["output"]
rule pre_proc:
params:
opt_exec="echo %RNDM:10 > seed.txt",
opt_args="--outputs seed.txt --nJobs 2 --avoidVP"
output: f"{config['output']}.pre_proc"
shell: "prun"
# noinspection SmkUnrecognizedSection
rule main_hpo:
params:
opt_trainingDS=rules.pre_proc.output[0],
opt_args="--loadJson conf.json"
input: rules.pre_proc.output
output: f"{config['output']}.main_hpo"
when: str(param_of("opt_trainingDS"))
shell: "prun"
# noinspection SmkUnrecognizedSection
rule post_proc:
params:
opt_inDS=rules.main_hpo.output[0],
opt_exec="echo %IN > anal.txt",
opt_args="--outputs anal.txt --nFilesPerJob 100 --avoidVP"
input: rules.main_hpo.output
output: config["output"]
when: str(param_of("opt_inDS"))
shell: "prun"
Loops in workflows
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection SmkRuleRedeclaration
rule all:
input: config["output"]
rule work_start:
params:
opt_exec="echo %RNDM:10 > seed.txt",
opt_args="--outputs seed.txt --nJobs 2 --avoidVP"
output: f"{config['output']}.work_start"
shell: "prun"
# noinspection SmkUnrecognizedSection, SmkRuleRedeclaration
rule work_loop:
params:
dataset=rules.work_start.output[0]
input: rules.work_start.output
loop: True
output: f'{config["output"]}.work_loop'
shell: "../loop_body/Snakefile"
rule work_end:
params:
opt_inDS=rules.work_loop.output[0],
opt_exec="echo %IN > results.root",
opt_args="--outputs results.root --forceStaged"
input: rules.work_loop.output
output: config["output"]
shell: "prun"
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of, param_exp
# noinspection SmkRuleRedeclaration
rule all:
params:
dataset=None,
param_xxx=123,
param_yyy=0.456
input: config["output"]
rule inner_work_top:
params:
opt_inDS=param_of('dataset',source=rules.all),
opt_exec=param_exp("echo %IN %{xxx} %{i} > seed.txt"),
opt_args="--outputs seed.txt --avoidVP"
output: f"{config['output']}.inner_work_top"
shell: "prun"
rule inner_work_bottom:
params:
opt_inDS=rules.inner_work_top.output[0],
opt_exec=param_exp("echo %IN %{yyy} > results.root"),
opt_args="--outputs results.root --forceStaged"
input: rules.inner_work_top.output
output: f"{config['output']}.inner_work_bottom"
shell: "prun"
# noinspection SmkRuleRedeclaration
rule checkpoint:
params:
opt_inDS=[rules.inner_work_top.output[0], rules.inner_work_bottom.output[0]],
opt_exec=param_exp("echo %{DS1} %{DS2} aaaa; echo '{\"x\": 456, \"to_continue\": false}' > results.json")
input: rules.inner_work_top.output,rules.inner_work_bottom.output
output: config["output"]
shell: "junction"
loop keyword was added as Snakemake language extension to define step as a loop.
Loop + scatter
Running multiple loops in parallel
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of
# noinspection SmkRuleRedeclaration
rule all:
params:
signals=[]
input: config["output"]
# noinspection SmkUnrecognizedSection, SmkRuleRedeclaration
rule work_loop:
params:
dataset=param_of('signals',source=rules.all)
scatter: [param_of('dataset')]
loop: True
output: f'{config["output"]}.work_loop'
shell: "../loop_body/Snakefile"
# noinspection SmkRuleRedeclaration
rule merge:
params:
opt_inDS=rules.work_loop.output[0],
opt_exec="echo %IN > results.root",
opt_args="--outputs results.root --forceStaged"
output: config["output"]
shell: "prun"
A sub-workflow filename should be specified in shell keyword.
Parallel execution of multiple sub-workflows in a single loop
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of
# noinspection SmkRuleRedeclaration
rule all:
params:
signal=None
input: config["output"]
# noinspection SmkUnrecognizedSection
rule seq_loop:
params:
dataset=param_of('signal',source=rules.all)
loop: True
output: config["output"]
shell: "../scatter_body/Snakefile"
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of, param_exp
# noinspection SmkRuleRedeclaration
rule all:
params:
dataset=None,
param_xxx=[123, 456],
param_yyy=[0.456, 0.866]
input: config["output"]
# noinspection SmkRuleRedeclaration, SmkUnrecognizedSection
rule parallel_work:
params:
dataset=param_of('dataset',source=rules.all),
param_sliced_x=param_of('param_xxx',source=rules.all),
param_sliced_y=param_of('param_yyy',source=rules.all)
scatter: [param_of('param_sliced_x'), param_of('param_sliced_y')]
output: f'{config["output"]}.parallel_work'
shell: "../loop_main/Snakefile"
# noinspection SmkRuleRedeclaration
rule checkpoint:
params:
opt_inDS=rules.parallel_work.output[0],
opt_exec=param_exp(
"echo '{\"xxx\": [345, 678], \"yyy\": [0.321, 0.567], \"to_continue\": false}' > results.json"
)
output: config["output"]
input: rules.parallel_work.output
shell: "junction"
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_of
# noinspection SmkRuleRedeclaration
rule all:
params:
dataset=None,
param_sliced_x=None,
param_sliced_y=None
input: config["output"]
rule core:
params:
opt_inDS=param_of("dataset",source=rules.all),
opt_exec=lambda wildcards: "echo %IN %{sliced_x} %{sliced_y} > seed.txt",
opt_args="--outputs seed.txt --avoidVP"
output: config["output"]
shell: "prun"
Using REANA
# noinspection PyUnresolvedReferences
configfile: "../config.json"
# noinspection PyUnresolvedReferences
from pandaserver.workflow.snakeparser.utils import param_exp
# noinspection SmkRuleRedeclaration
rule all:
input: config["output"]
rule ain:
params:
opt_exec="echo %RNDM:10 > seed_ain.txt",
opt_args="--outputs seed_ain.txt --avoidVP --nJobs 2"
output: f"{config['output']}.ain"
shell: "prun"
rule twai:
params:
opt_exec="echo %RNDM:10 > seed_twai.txt",
opt_args="--outputs seed_twai.txt --avoidVP --nJobs 3"
output: f"{config['output']}.twai"
shell: "prun"
rule drai:
params:
opt_inDS=[rules.ain.output[0], rules.twai.output[0]],
opt_exec=param_exp("echo %{DS1} %{DS2} > results.root"),
opt_args="--outputs results.root",
opt_containerImage="docker://busybox"
input: rules.ain.output,rules.twai.output
output: config["output"]
shell: "reana"
Debugging locally
Workflow descriptions can be error-prone. It is better to check workflow descriptions before submitting them.
pchain has the --check option to verify the workflow description locally.
You just need to add the --check option when running pchain.
For example,
pchain --cwl test.cwl --yaml dummy.yaml --outDS user.<your_nickname>.blah --check
which should give a message like
INFO : uploading workflow sandbox
INFO : check workflow user.tmaeno.c63e2e54-df9e-402a-8d7b-293b587c4559
INFO : messages from the server
internally converted as follows
ID:0 Name:top Type:prun
Parent:
Input:
opt_args: --outputs seed.txt --nJobs 2 --avoidVP
opt_exec: echo %RNDM:10 > seed.txt
Output:
...
INFO : Successfully verified workflow description
Your workflow description is sent to the server to check
whether the options in opt_args are correct, dependencies among steps are valid,
and input and output data are properly resolved.
Monitoring
Bookkeeping
pbook provides the following commands for workflow bookkeeping
show_workflow
kill_workflow
retry_workflow
finish_workflow
pause_workflow
resume_workflow
Please refer to the pbook documentation for their details.