Running Athena analysis
This page explains how to run Athena analysis on PanDA.
Preparation
First you need to setup the runtime environment of Athena which you want to use on PanDA. Here is an example with AnalysisBase,21.2.40
export ATLAS_LOCAL_ROOT_BASE=/cvmfs/atlas.cern.ch/repo/ATLASLocalRootBase
alias setupATLAS='source ${ATLAS_LOCAL_ROOT_BASE}/user/atlasLocalSetup.sh'
setupATLAS
asetup AnalysisBase,21.2.40,here
Checkout packages, add modifications, and build them if necessary. Make sure that your Athena job runs properly.
Then you need to setup panda-client.
lsetup panda
Once everything is done you can try
pathena --helpGroup ALL
to see all available options.
How to run
Running jobOptions
When you run Athena with
athena jobO_1.py jobO_2.py jobO_3.py
all you need is
pathena jobO_1.py jobO_2.py jobO_3.py <--inDS inputDataset> --outDS outputDataset
where --inDS takes a dataset, a dataset container name, or a comma-concatenated list
of input dataset/container names if the job read input data,
while --outDS takes the base name of output dataset containers.
pathena parses jobOptions to define output types, collects various environment variables,
makes a relocatable sandbox file from your local built packages to recreate your local runtime environment
on remote resources, and submits a task together with those materials to PanDA.
EventSelector.InputCollections is automatically set to read files from --inDS so that
you don’t have to change anything in your jobOptions files.
One output dataset container is created for each output type and is registered in rucio.
If you want to monitor your task, see Monitoring.
Once your task is done you will get an email notification, then you can download output
files using rucio client. Your output files are available for 30 days.
If you want to retry your task, see Bookkeeping.
Running transformations
The --trf option of pathena allows users to run official transformations such as Reco_tf.py
with customized packages. The option takes an execution string which can be created as follows:
If you locally run a trf like
Reco_tf.py inputAODFile=AOD.493610._000001.pool.root.1 outputNTUP_SUSYFile=my.NTUP.root"
replace some parameters with %XYZ using the following table
Parameter |
Placeholder |
|---|---|
Input |
%IN |
Cavern Input |
%CAVIN |
Minimumbias Input |
%MININ |
Low pT Minimumbias Input |
%LOMBIN |
High pT Minimumbias Input |
%HIMBIN |
BeamHalo Input |
%BHIN |
BeamGas Input |
%BGIN |
Output |
%OUT + suffix (e.g., %OUT.ESD.pool.root) |
MaxEvents |
%MAXEVENTS |
SkipEvents |
%SKIPEVENTS |
FirstEvent |
%FIRSTEVENT:basenumber (e.g., %FIRSTEVENT:100, the base number is given to the first job and it will be incremented per job) |
DBRelease or CDRelease |
%DB:DatasetName:FileName (e.g., %DB:ddo.000001.Atlas.Ideal.DBRelease.v050101:DBRelease-5.1.1.tar.gz. %DB:LATEST if you use the latest DBR). Note that if your trf uses named parameters (e.g., DBRelease=DBRelease-5.1.1.tar.gz) you will need DBRelease=%DB:DatasetName:FileName (e.g., DBRelease=%DB:ddo.000001.Atlas.Ideal.DBRelease.v050101:DBRelease-5.1.1.tar.gz) |
Random seed |
%RNDM:basenumber (e.g., %RNDM:100, this will be incremented per job) |
Then you just need to give the string to --trf, e.g.
pathena --trf "Reco_trf.py inputAODFile=%IN outputNTUP_SUSYFile=%OUT.NTUP.root" --inDS ... --outDS ...
When your trf doesn’t take an input (e.g., evgen), use the --split option to specify how many jobs you need.
%SKIPEVENTS may be needed if you use the --nEventsPerJob or --nEventsPerFile options of pathena.
Otherwise, some jobs will run on the same event range in the same input file.
Note that you may need to explicitly specify maxEvents=XYZ or something in the execution string to set the number
of events processed in each job, since the value of --nEventsPerJob or --nEventsPerFile
is used only to split files, but is not appended to the execution string.
Otherwise, each job will process all events in the input file.
pathena doesn’t interpret the argument for the –trf option although it replaces %XYZ.
It is user’s responsibility to consistently specify pathena options and the execution string.
If you want to add parameters to the transformation that are not listed above, just add them to the execution string.
pathena doesn’t replace anything except %XYZ, but it passes these parameters along to the transformation
just the same.
Running multiple transformations
One can run multiple transformations in a job by using semicolons in the --trf option like
pathena --trf "trf1.py ...; trf2.py ...; trf3.py ..." ...
Here is an example to run simul+digi;
pathena --trf "AtlasG4_trf.py inputEvgenFile=%IN outputHitsFile=tmp.HITS.pool.root maxEvents=10 skipEvents=0 randomSeed=%RNDM geometryVersion=ATLAS-GEO-16-00-00 conditionsTag=OFLCOND-SDR-BS7T-04-00; Digi_trf.py inputHitsFile=tmp.HITS.pool.root outputRDOFile=%OUT.RDO.pool.root maxEvents=-1 skipEvents=0 geometryVersion=ATLAS-GEO-16-00-00 conditionsTag=OFLCOND-SDR-BS7T-04-00" --inDS ...
where AtlasG4_trf.py produces a HITS file (tmp.HITS.pool.root) which is used as an input by Digi_trf.py to produce RDO. In this case, only RDO is added to the output dataset since only RDO has the %OUT prefix (i.e. %OUT.RDO.pool.root).
If you want to have HITS and RDO in the output dataset the above will be
pathena --trf "AtlasG4_trf.py inputEvgenFile=%IN outputHitsFile=%OUT.HITS.pool.root maxEvents=10 skipEvents=0 randomSeed=%RNDM geometryVersion=ATLAS-GEO-16-00-00 conditionsTag=OFLCOND-SDR-BS7T-04-00; Digi_trf.py inputHitsFile=%OUT.HITS.pool.root outputRDOFile=%OUT.RDO.pool.root maxEvents=-1 skipEvents=0 geometryVersion=ATLAS-GEO-16-00-00 conditionsTag=OFLCOND-SDR-BS7T-04-00" --inDS ...
Note that both AtlasG4_trf.py and Digi_trf.py take %OUT.RDO.pool.root as a parameter. AtlasG4_trf.py uses it as an output filename while Digi_trf.py uses it as an input filename.
Running arbitrary executables in the Athena runtime environment
If you want to run arbitrary executables available in the Athena runtime environment, you can use the --trf option
as it is essentially equivalent to --exec plus --useAthenaPackages of the prun command.
The --trf option skips the job options parsing and auto job configuration, so you need to specify
parameters for the executables using placeholders, listed at the above section,
such as %IN and %NEVENTS, and output files using %OUT or --extOutFile.
Note that Athena with ComponentAccumulator-based configuration behaves more like transformation than traditional
one with jobOptions, so it is easier to use the --trf option in that case.
Here are a few examples:
pathena --trf "python -m AthExHelloWorld.HelloWorldConfig --filesInput=%IN --evtMax=%MAXEVENTS --profile-python=%OUT.txt" --nEventsPerJob=3 ...
where input filenames and the number of events per job are set by %IN and %MAXEVENTS respectively, which are
automatically replaced with actual values when the job runs. The output file is set by %OUT.txt, so the
python code profiles are uploaded as output files.
If you are using the --CA option of athena.py,
get_files AthExHelloWorld/HelloWorldConfig.py
pathena --trf "athena.py --CA HelloWorldConfig.py --evtMax=%MAXEVENTS --filesInput=%IN --profile-python=aaa.txt" --nEventsPerJob=3 --extOutFile=aaa.txt ...
where the --extOutFile option is used to specify the output file.
Or if it is executed as a script,
get_files AthExHelloWorld/HelloWorldConfig.py
printf '%s\n%s\n' '#!/usr/bin/env python' "$(cat HelloWorldConfig.py)" > HelloWorldConfig.py
chmod +x HelloWorldConfig.py
pathena --trf "./HelloWorldConfig.py --evtMax=%MAXEVENTS --filesInput=%IN --profile-python=%OUT.txt" --nEventsPerJob=3 ...
Reading particular events for data
You can specify a run/event list as an input. First you need to prepare a list of runs/events of interest. You may get a list by analysing D3PD, browsing event display, using ELSSI, and so on. A list looks like
$ cat rrr.txt
154514 21179
154514 29736
154558 448080
where each line contains a run number and an event number. Then, e.g.,
pathena AnalysisSkeleton_topOptions.py --eventPickEvtList rrr.txt --eventPickDataType AOD \
--eventPickStreamName physics_CosmicCaloEM --outDS user...
where events in the input file are internally converted to AOD (specifid by --eventPickDataType) with
the physics_CosmicCaloEM stream (specified by --eventPickStreamName).
Your jobO is dynamically configured to use event selection, so you don’t need to change your jobO.
In principle, you can run any arbitrary jobO.
FAQ
Contact
We have one egroup and one JIRA. Please submit all your help requests to hn-atlas-dist-analysis-help@cern.ch which is maintained by AtlasDAST.
How PanDA chooses sites where jobs run?
PanDA chooses sites using the following information;
input data locality
the number of jobs in activated/defined/running state (site occupancy rate)
the average number of CPUs per worker node at each site
the number of active or available worker nodes
pilot rate for last 3 hours. If no pilots, the site is skipped
available disk space in SE
Atlas release/cache matching
site statue
and then calculate the weight for each site using the following formula.
where
W: Weight at the site
G: The number of available worker nodes which have sent getJob requests for last 3 hours
U: The number of active worker nodes which have sent updateJob requests for last 3 hours
R: The maximum number of running jobs in last 24 hours
D: The number of defined jobs
A: The number of activated or starting jobs
T: The number of assigned jobs which are transferring input files to the site
X: Weight factor based on data availability. When input file transfer is disabled, X=1 if input data is locally available, otherwise X=0. When input file transfer is enabled, X=1+(total size of input files on DISK)/10GB if files are available on DISK, X=1+(total size of input files on TAPE)/1000GB if files are available on TAPE, X=1 otherwise
What are buildJob and runJob?
Once PanDA chooses sites to run jobs the relocatable sandbox file is sent to the sites. One buildJob is created at each site to upload the sandbox file to the local storage at the site. The completion of buidJob triggers a bunch of runJobs. Each runJob retrieves the sandbox file to run Athena.
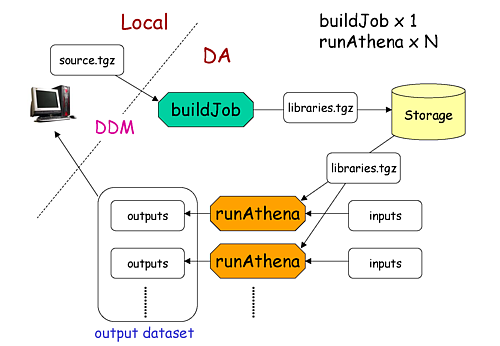
It is possible to skip buildJob using --noBuild. In this case runJobs retrive the sandbox file from
a web service but the size of the sandbox file is limted to 50MB.
How job priorities are calculated?
Job priorities are calculated for each user by using the following formula. When a user submits a task composed of M jobs,
where Priority(n) is the priority of n-th job (0≤n<M), and the total number of the user’s active jobs in the whole system.
For example, if a fresh user submits a task composed of 100 jobs, the first 5 jobs have priority=1000 while the last 5 jobs have priority=981. The idea of this gradual decrease is to prevent huge tasks from occupying the whole CPU slots. When another fresh user submits a job with 10 jobs, these jobs have priority=1000,999 so that they will be executed as soon as CPU becomes available even if other users have already queued many jobs. Priorities for waiting jobs in the queue are recalculated every 20 minutes. Even if some jobs have very low priorities at the submission time their priorities are increased periodically so that they are executed before they expire.
If the user submits jobs with the --voms and --official options to produce group datasets, those jobs
are regarded as group jobs. Priorities are calculated per group separately from the user who submitted, so group
jobs don’t reduce priorities of normal jobs which are submitted by the same user without those options.
There are a few kinds of jobs which have higher priorities, such as merge jobs (5000) and HummerCloud jobs (4000), since they have to be processed quickly.
How can I send jobs to the site which holds the most number of input files?
You can send jobs to a specific site using --site, but the option is not recommended,
since Jobs should be automatically sent to proper sites.
Why is my job pending in activated/defined state?
Jobs are in the activated state until CPU resources become available at the site. If the site is busy your jobs will have to wait so long. runJobs are in defined state until corresponding buildJobs have finished.
How jobs get reassigned to other sites? Why were my jobs reassigned by JEDI?
Jobs are internally reassigned to another site at most 3 times, when
they are waiting for 24 hours.
HammerCloud set sites to the test or offline mode 3 hours ago
The algorithm for site selection is the same as normal brokerage described in the above section. Old jobs are closed. When a new site is not found, jobs will stay at the original site.
How to debug failed jobs
You can see the error description in Monitoring.
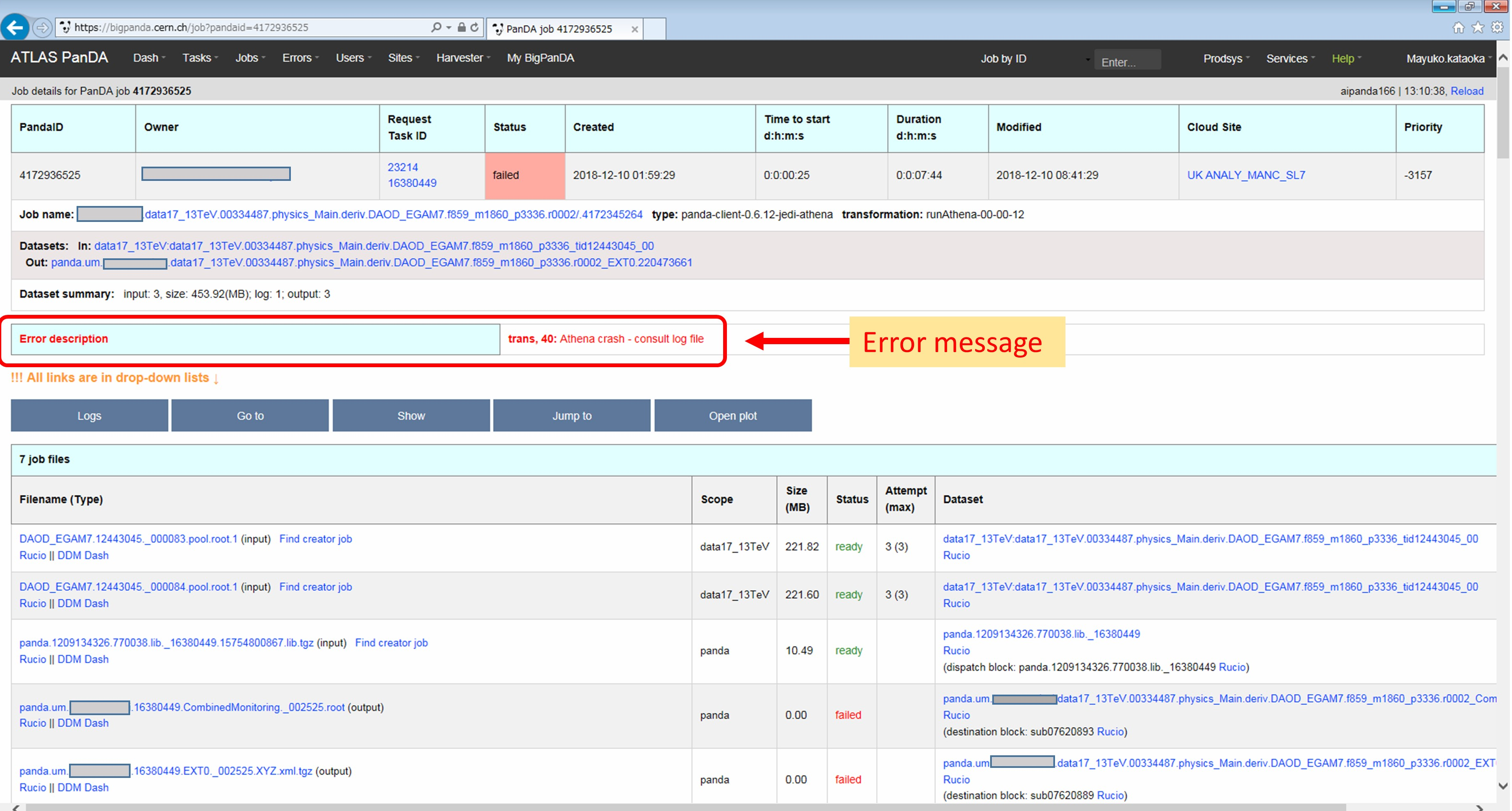
When transExitCode is not zero, the job failed with an Athena problem. You may want to see log files. You can browse the log files following links “Logs” → “Log files”.
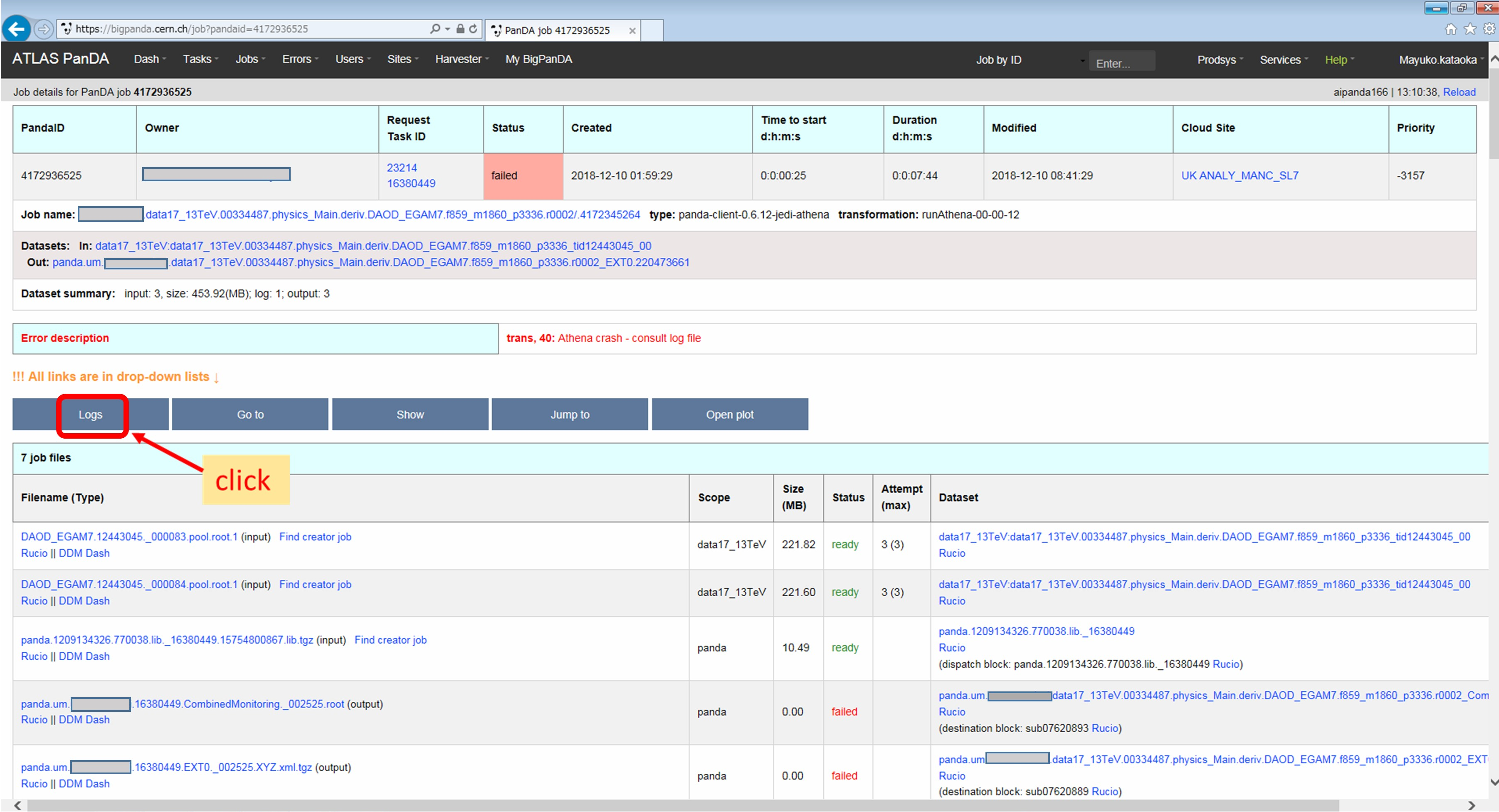
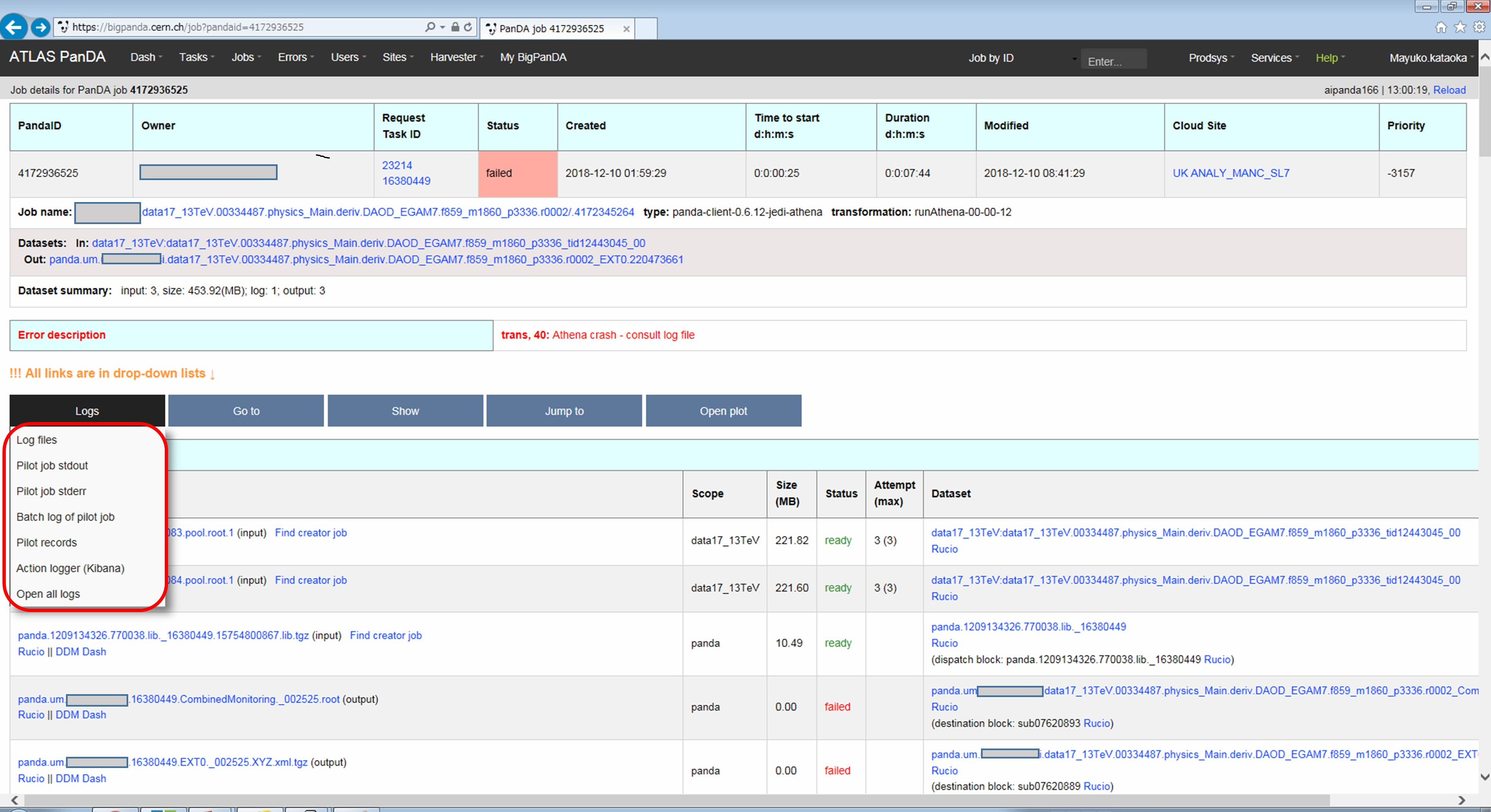
Now you find various log files shown in the page.
E.g., there should be payload.stdout for stdout and payload.stderr for stderr, where you may get some clues.

Note that some filed jobs don’t have log files. This typically happens when jobs are killed by the batch system before uploading log files.
Why did my jobs crash with “sh: line 1: XYZ Killed”?
sh: line 1: 13955 Killed athena.py -s ...
If you see something like the above message in the log file, perhaps your jobs were killed by
the batch system due to huge memory consumption. You may explicitly reduce the number of input files per job
using --nFilesPerJob if memory consumption scales with the number of files. However, not set a very small
number to --nFilesPerJob. If your jobs are very short the system will automatically ignore --nFilesPerJob
since too many short jobs kill the system.
Why were my jobs closed ? What does ‘upstream job failed’ mean?
If a buildJob fails, corresponding runJobs will get closed.
Are there any small samples to test my job before run it on the whole dataset?
You can limit the number of files to be used in --inDS by using --nFiles.
What is the meaning of the ‘lost heartbeat’ error?
Each job sends heartbeat messages every 30 min to indicate it is alive. If there is no heartbeat message for 6 hours, the job gets killed. The error typically happens when the job died due to temporary troubles in the backend batch system or network. Generally jobs are automatically retried and the next attempts succeed.
What is the meaning of the ‘Looping job killed by pilot’ error?
If a job doesn’t update output files for 2 hours, it will be killed. This protection is intended to kill dead-locked jobs or infinite-looping jobs. If your job doesn’t update output files very frequently (e.g., some heavy-ion job takes several hours to process one event) you can relax this limit by using the –maxCpuCount option. However, sometimes even normal jobs get killed due to this protection. When the storage element has a problem, jobs cannot copy input files to run Athena and of course cannot update output files. When you think that your job was killed due to an storage problem, you may report to DAST. Then shift people and the SE admin will take care of it.
I want to process the whole dataset with N events per job. (integer N>0)
Use --nEventsPerJob that splits input files per nEventsPerJob and generates a job for each input chunk.
Note that the value of --nEventsPerJob
is mainly used to split files, and the value is automatically propagated to the payload
as theApp.EvtMax when you run normal jobOptions files, i.e. --trf is not used.
If the number of events in the payload is configured not through
theApp.EvtMax or --trf is used, specify maxEvents=XYZ or something consistently
in your jobOptions file, or the execution string if the --trf option is used.
--nEventsPerJob can be used even if your jobs don’t take input files. In this case, it just sets
theApp.EvtMax accordingly.
I want to launch M jobs, each with N events per job
You can use the following command:
pathena --split M --nEventsPerJob N .....
Note that --nFilesPerJob and --nEventsPerJob can not be defined simultaneously, pathena
will exit with an error at startup. Please define only one or another.
Expected output file does not exist
Perhaps the output stream is defined in somewhere in your jobOs, but nothing uses it. In this case,
Athena doesn’t produce the file. The solutions could be to modify your jobO or to use the --supStream option.
E.g., –supStream hist1 will disable user.aho.TestDataSet1.hist1._00001.root.
How to make group datasets
Use --official and --voms options.
pathena --official --voms atlas:/atlas/groupName/Role=production --outDS group.groupName.[otherFields].dataType.Version ...
where groupName for SUSY is phys-susy, for example.
See the document ATL-GEN-INT-2007-001 for dataset naming convention.
The group name needs to be officially approved and registered in ATLAS VOMS. Note that you need to have the production role
for the group to produce group-defined datasets. If not, please request it in the ATLAS VO registration page.
If you submit tasks with the --voms option, jobs are counted in the group’s quota.
How do I blacklist sites when submitting tasks?
Use --excludedSite. However, this option is not recommend since that would skew workload distrubution in the
whole system and decrease the system throughput.
How do I get the output dataset at my favorite destination automatically
When --destSE option is used, output files are automatically aggregated to a RSE. e.g.,
pathena --destSE LIP-LISBON_LOCALGROUPDISK ...
Generally LOCALGROUPDISK (long term storage) or SCRATCHDISK (short term storage) can be used. You can check permission in each RSE page in CRIC. For example, only /atlas/pt users are allowed to write to LIP-LISBON_LOCALGROUPDISK, so if you don’t belong to the pt group the above example will fail and you will have to choose another RSE.
pathena failed due to “ERROR : Could not parse jobOptions”
The error message would be something like:
ABC/XYZ_LoadTools.py", line 65, in <module>
input_items = pf.extract_items(pool_file=
svcMgr.EventSelector.InputCollections[0])
IndexError: list index out of range
ERROR : Could not parse jobOptions
First, make sure that you jobOptions work on your local computer without any changes. Basically pathena doesn’t work if Athena locally fails with the jobO.
For example, if it fails in InputFilePeeker, the solution is to have something like
svcMgr.EventSelector.InputCollections=["/somedir/mc08.108160.AlpgenJimmyZtautauNp0VBFCut.recon.ESD.e414_s495_r635_tid070252/ESD.070252._000001.pool.root.1"]
in your jobO, where the input file must be valid (i.e. can be accessed from your local computer).
Note that input parameter (essentially EventSelector.InputCollections and AthenaCommonFlags.FilesInput)
will be automatically overwritten to read input files in --inDS.
The local file doesn’t have to be from --inDS as long as
the data type, such as AOD,ESD,RAW…, is identical.
My task got exhausted due to over_cpu_consumption
This message means that jobs may have abused more CPU cores than allocation since the CPU time of your jobs was longer
than their total execution time.
Jobs can use more than
one CPU core even if they run on single-core queues since they physically run on multi-core CPUs.
This typically happens when your application internally spawns multiple threads/processes
and spreads over multiple CPU cores.
If this is the case, it would help send jobs to multi-core queues using the --nCore option.